Differential proteomics allows the comparison of big sets of proteins originating from various biological sources, like treated and untreated cells, or differential bacterial strains. Small differences in the big number of components can be discovered by differential proteomics.
Lumiprobe offers Cyanine2, Cyanine3, and Cyanine5 minimal dyes which are spectrally distinct, but mobility-matched. Thus, proteins labeled with these minimal dyes via NHS ester chemistry co-migrate in gel-electrophoresis. So, lysines are the predominant sites of labeling.
The best practice of 2D proteomics makes use of an internal pooled control sample which is essentially a mixture of both proteomes labeled with one of the dyes. Cyanine2 should be used for it. The other two dyes, Cyanine3, and Cyanine5 are interchangeable for all experiments.
Fluorescent minimal dyes come pre-measured in 5 nmol, 10 nmol, and 25 nmol packagings. Dyes are lyophilized to prolong shelf life. Each package should be re-constituted with DMF prior to use. DMF quality is essential for the experiments, and product shelf life. DMF (dimethylformamide), labeling grade is available for purchase from our catalog list.
Protein mixture preparation is the most variable and most demanding part of the assay. It is outside the scope of the official protocol because different samples require very different lysis conditions to obtain a mixture of proteins suitable for the assay. Essentially, both samples being compared should be lysed in identical conditions to achieve meaningful results. Generally, if the experiment requires lysis, CHAPS-based lysis solutions are recommended.
After the preparation of both protein mixtures being compared, the following protocol can be used to achieve labeling.
1) Prepare 1 mM stock solution of each dye by adding 1 uL of DMF per 1 nmol of each dye. These solutions are stable for three months at 20°C. To ensure shelf life, desiccate, and completely unfreeze the tubes before opening, purge them with inert gas when possible before closing
2) Adjust protein mixtures pH to 8.5 by using either amine-free buffer (NaHCO3, acetate) or Tris buffer. In separate vials, prepare three labeling reactions: one untreated, one treated, and one mix of both (pooled internal control). Protein concentration should ideally be 5-10 mg/mL, but as low as 1 mg/mL can be used. Take 50 ug of protein per reaction.
3) Prepare working solutions of dyes by taking aliquots of 1 mM stock solution and diluting with DMF to 0.4 mM.
4) Add 1 uL of working dye solution to the reaction mixture: Cyanine3 for untreated, Cyanine5 for treated (or vice versa), and Cyanine2 for pooled internal control. Mix each reaction by pipetting it in and out, and leave for 30 min in the dark.
5) Stop the reactions by adding 1 uL of 10 mM lysine to each solution
6) Pool the three samples, and run the 2D gel.
7) Analyze with any imager capable of detecting Cyanine2, Cyanine3, and Cyanine5. Protein spots can be cut, and analyzed by mass spectrometry.
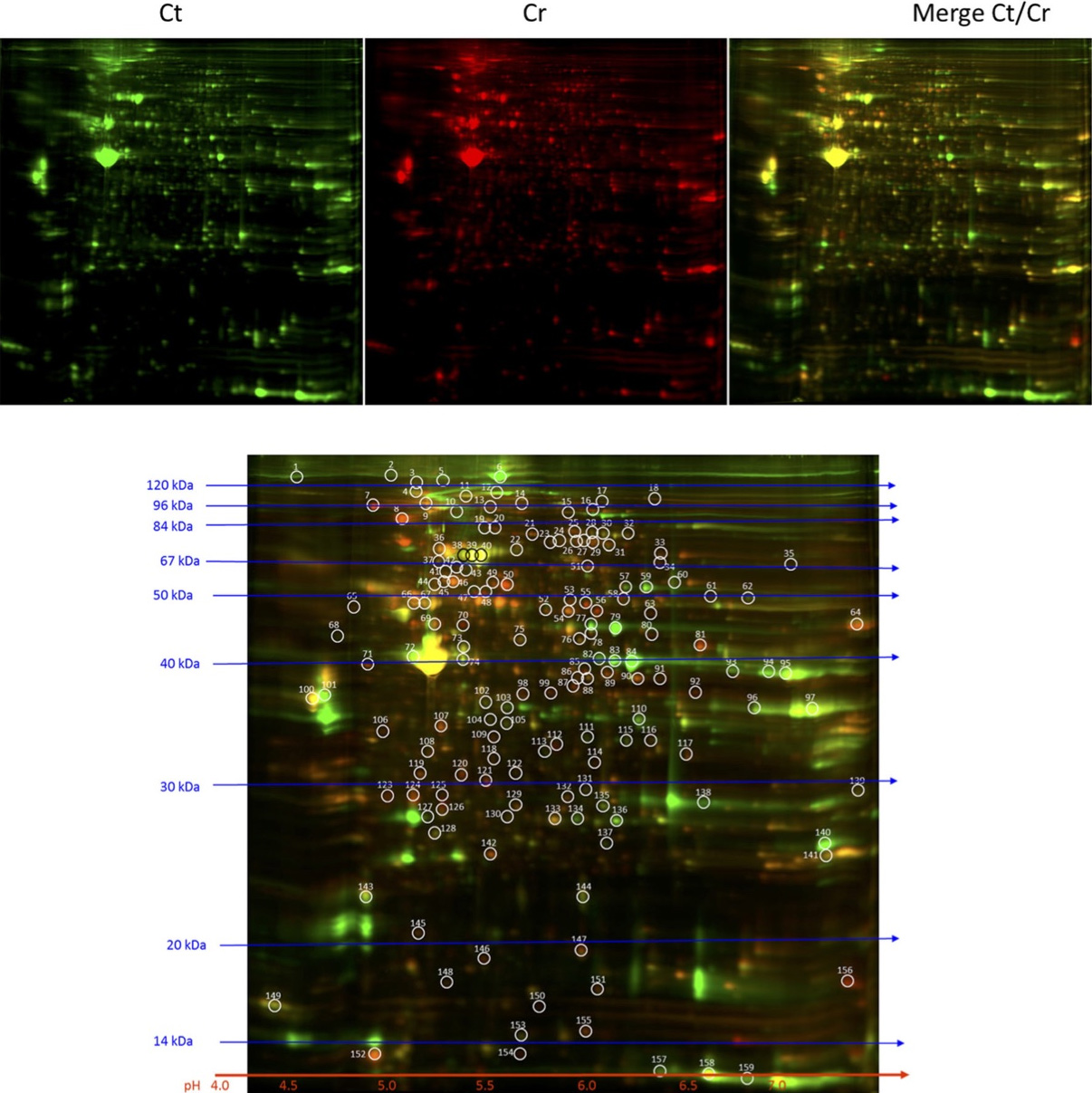
Below is the example of a 2D-DIGE image of the detection of differentially expressed proteins in Cr(VI)-transformed Beas-2B cells by 2D-DIGE labeled using our fluorescent minimal dyes (Lu et al.: 10.1007/s12011-017-1222-9):