dsGreen can be used as a dye for quantitative real-time PCR (qPCR). dsGreen selectively binds to dsDNA and forms a dsDNA–dye complex. The complex of the dye with double-stranded DNA provides fluorescence emission with a peak at 524 nm, so the fluorescence of the dsDNA–dye complex can be detected on the FAM channel on most common real-time PCR instruments (Green channel for Rotor-Gene® Q).
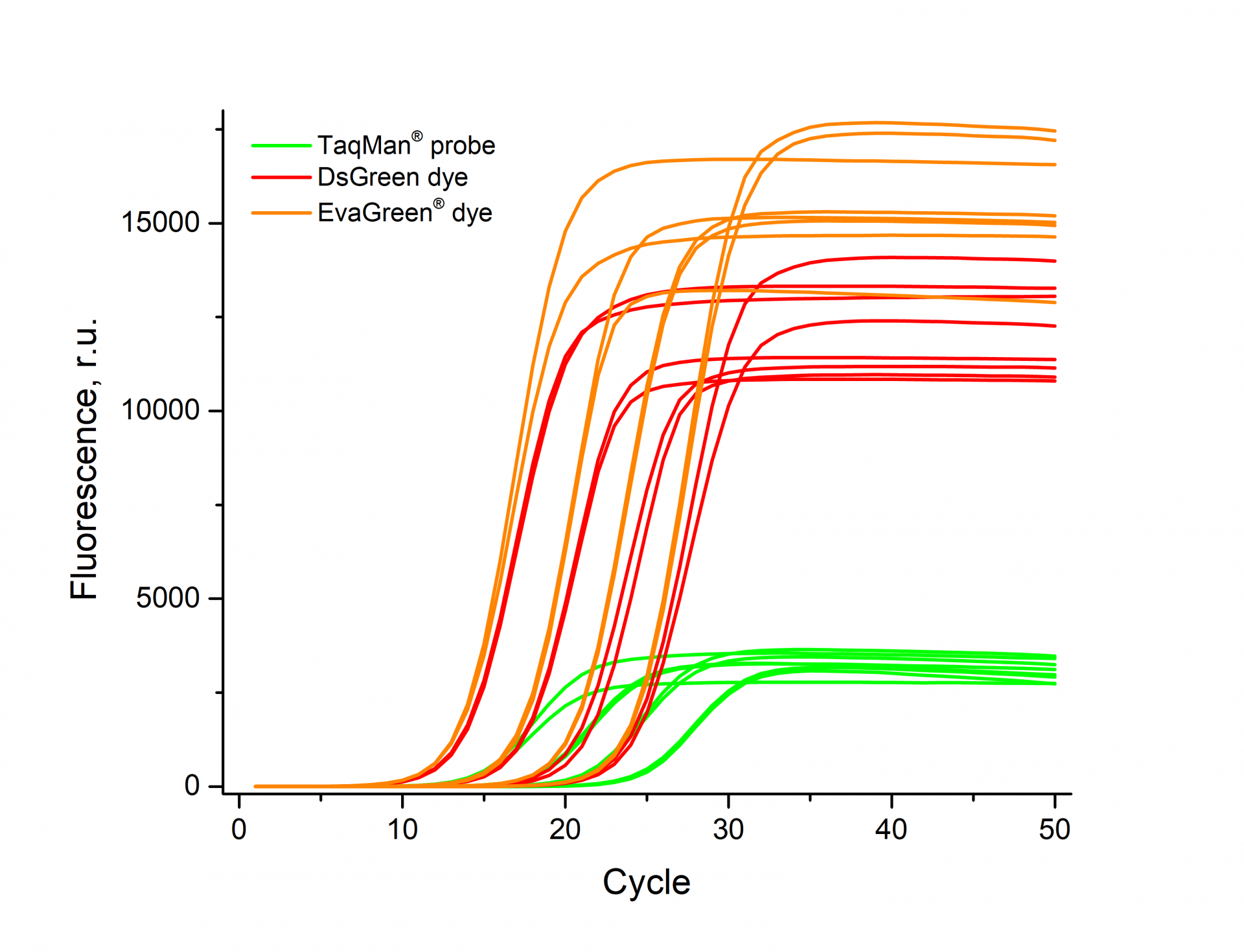
The concentration of dsGreen dye has been designed specially for qPCR, so Taq DNA polymerase is not inhibited by the dye* while maintaining high fluorescence intensity.
To set up a PCR reaction with dsGreen, you need Taq Polymerase with appropriate buffer containing Mg2+, dNTPs and primers, or PCR ready-to-use mix without any other fluorescent dyes. It is also recommended to use hot start Taq DNA Polymerase to reduce background from nonspecific amplification.
dsGreen is an intercalating dye, therefore it binds to any double-stranded DNA. For this reason special attention should be given to primer design and PCR optimization to avoid primer dimer formation and nonspecific amplification. To differentiate primer dimers from the desired specific PCR product, it is necessary to obtain the melting curve after PCR reaction has been completed. The melting temperature of amplicon is determined at the maximum of the negative first derivative of the melting-curve (use the HRM analysis (High Resolution Melting) if possible for your equipment). The melting temperature of primer dimers must be noticeably lower than those of specific PCR product.
The following protocol for PCR with dsGreen can be used when performing a PCR experiment.
*- was tested by determining the efficiency of PCR with dsGreen and comparing it with hydrolysis probes and EvaGreen® dye
Rotor-Gene® is a trademark of the QIAGEN Group
EvaGreen® is a trademark of Biotium
TaqMan® is a trademark of Roche Diagnostics, Inc.